On May 29, a research paper titled "Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity" was published online in the journal Nature. The study was conducted by the research team led by associate professor Jun-Jie Gogo Liu from the School of Life Sciences at Tsinghua University, in collaboration with the teams of associate professor Chunlai Chen from Tsinghua University and investigator Yang Bai from Peking University.
The CRISPR system is an adaptive immune system in prokaryotes that protects against foreign DNA invasion. Among its various types, Cas9 has been extensively studied and applied in genome editing due to its mechanism of RNA-guided DNA-nuclease. The II-C subtype of Cas9 exhibits high diversity, yet whether there are additional mechanisms in the CRISPR-Cas9 system to counteract viral immune evasion remains unclear.
By utilizing bioinformatics analysis, the researchers identified novel-associated genes (NAGs) enriched in the gene clusters of larger II-C type Cas9 proteins, suggesting their involvement in bacterial immunity mediated by Cas9. To further explore this relationship, the team developed a structural growth trajectory analysis (SGT analysis), which enabled them to predict and analyze the structures of 1,381 II-C type Cas9 proteins. They discovered that larger II-C Cas9 proteins show a trend of evolving through new functional domains acquirements, with NAGs enriched at the endpoints of these growth trajectories.
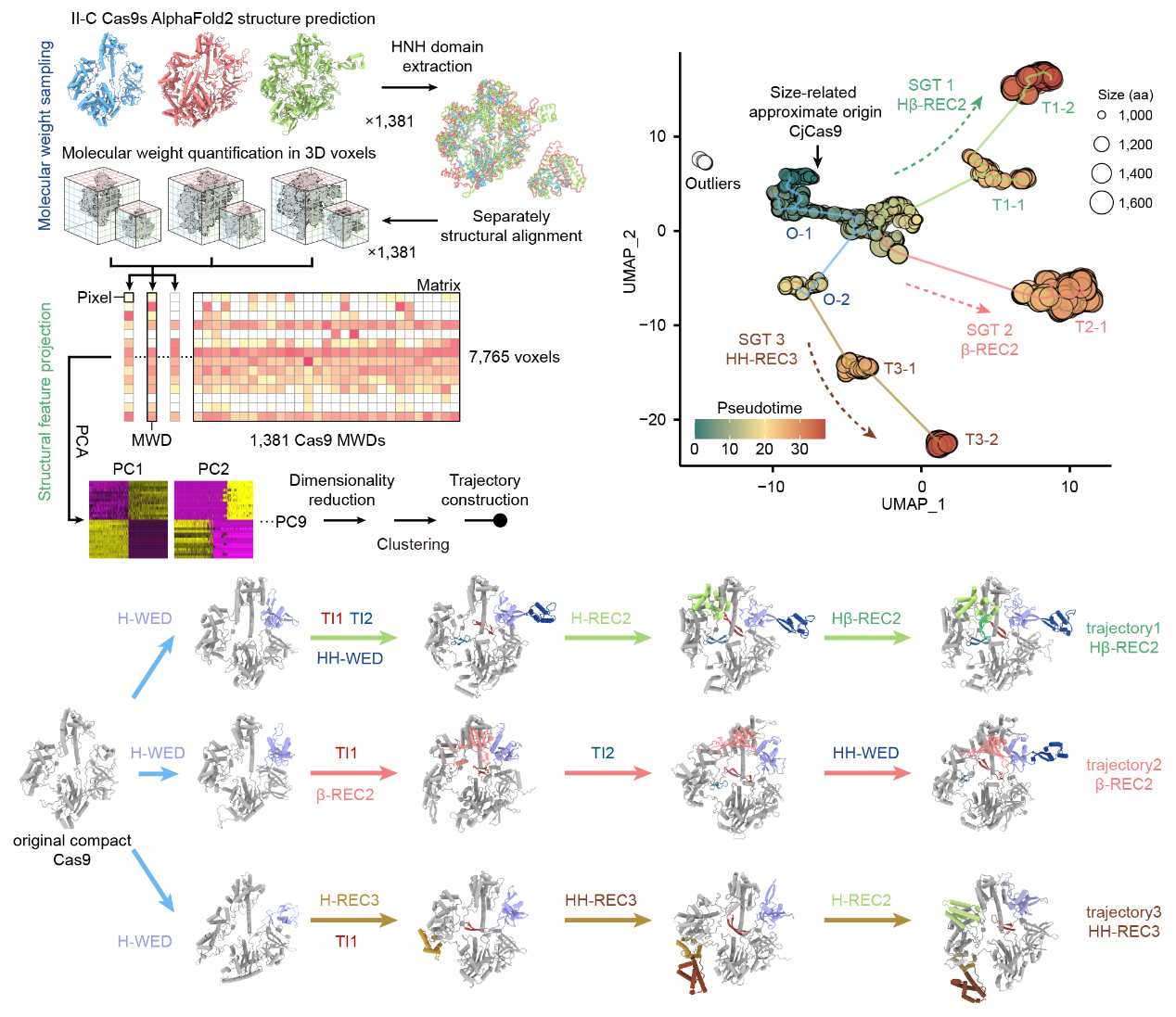
Structural growth trajectory analysis method (left) and growth trajectory diagram of II-C subtype Cas9 (right)
Biochemical experiments and cryo-electron microscopy revealed that the Cas9 protein from Chryseobacterium sp. (CbCas9) has developed a new β-REC2 domain that enhances Cas9 activity and a CTH domain that interacts with its associated gene-encoded PcrIIC1. The CTH domain allows the formation of a heterotetramer complex of two CbCas9 and two PcrIIC1 proteins.
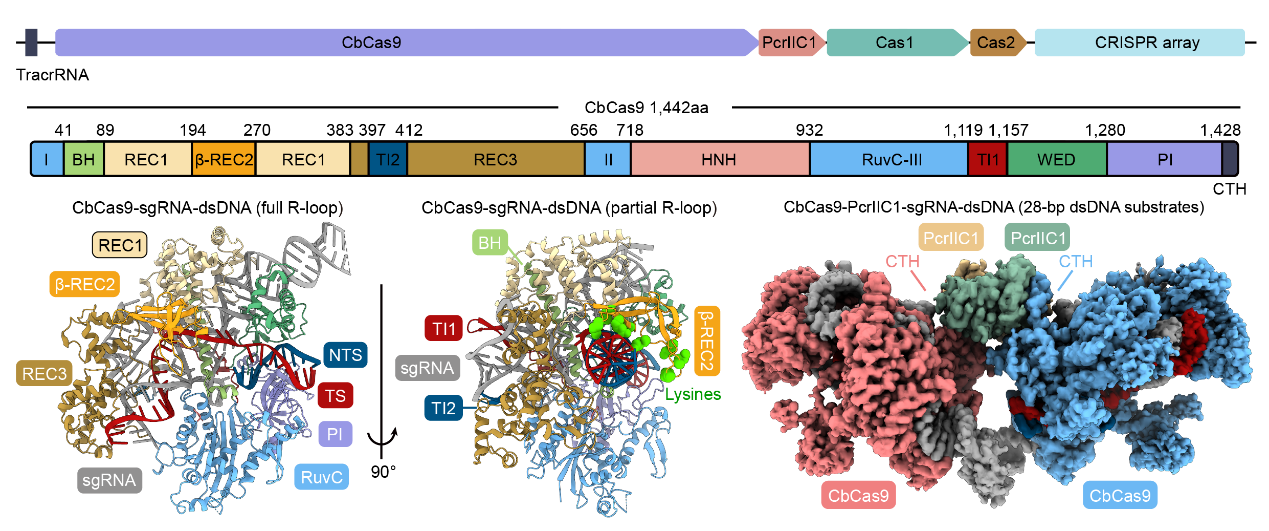
CbCas9 locus (top), CbCas9 effector protein structure (bottom left), and CbCas9-PcrIIC1 complex structure (bottom right)
The study demonstrated that the PcrIIC1 protein acts as a pro-CRISPR factor, significantly boosting the activity of the CbCas9 effectors. The CbCas9-PcrIIC1 complex exhibited enhanced DNA binding and cleavage activity, broader protospacer adjacent motif (PAM) compatibility, stronger DNA unwinding capability, and increased tolerance of target sequence mismatches.
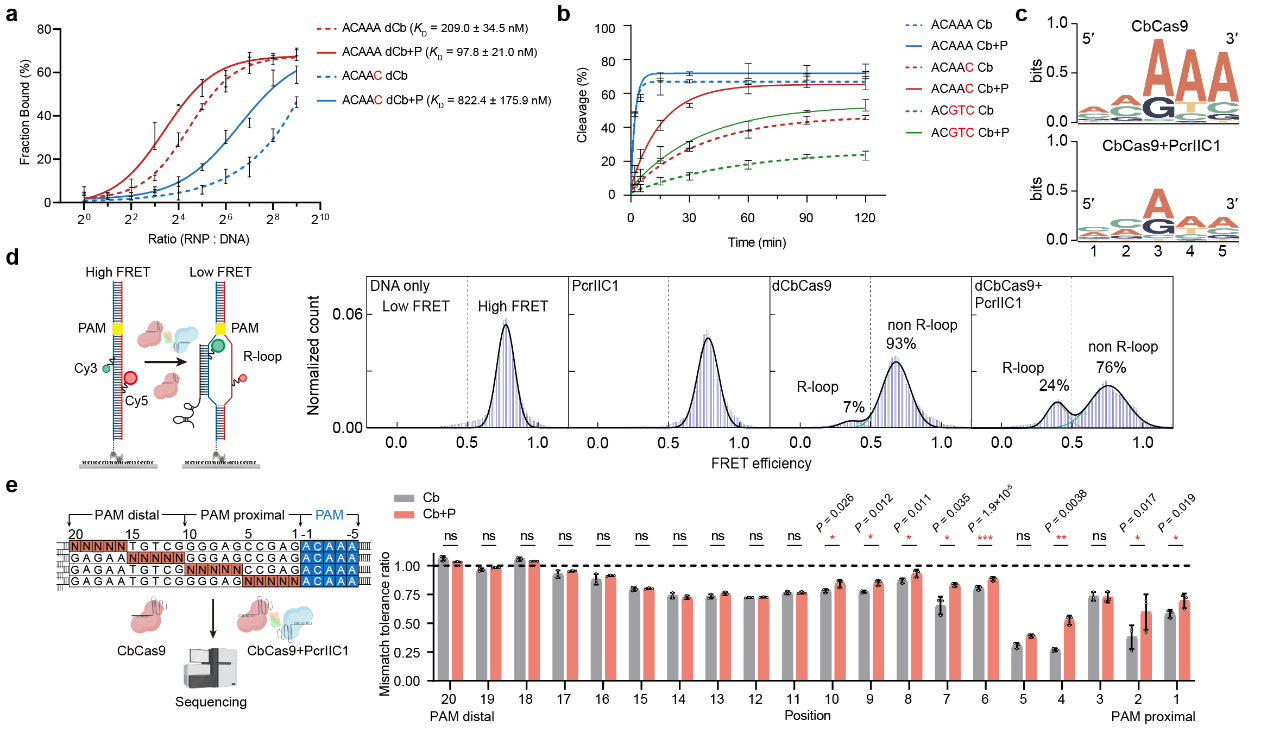
PcrIIC1 enhances CbCas9's DNA-binding (a), cleavage (b), PAM compatibility (c), DNA unwinding (d), and mismatch tolerance (e)
Structural analysis revealed that the enhanced activity of the CbCas9-PcrIIC1 complex is due to the stronger electrostatic interactions and the cooperative action of the two CbCas9 proteins binding to the same long-chain DNA. This synergistic effect facilitates DNA distortion and allosteric changes, enhancing the DNA interference capability of the complex. Additionally, the two CbCas9 proteins can symmetrically target sequences on both strands of one DNA target, improving the efficiency of genomic target search.
To validate the impact of PcrIIC1 on the anti-phage immunity of the CbCas9 system, the research teams performed the phage plaque assay in E. coli. The results showed that PcrIIC1 significantly enhances the phage resistance of the CbCas9 system. Disrupting the interaction between CbCas9 and PcrIIC1 led to the loss of enhanced immunity, indicating the critical role of the CbCas9-PcrIIC1 complex in boosting the CRISPR-Cas system's immune function.

PcrIIC1 significantly enhanced the bacterial immune activity of CbCas9 system
This study not only provides insights into the evolutionary trajectory of II-C subtype Cas9 but also identifies a novel pro-CRISPR factor that improve Cas9 activity through dimerization. These findings pave the way for developing more efficient gene editing tools based on pro-CRISPR factors.
Associate professor Jun-Jie Liu, investigator Yang Bai, and associate professor Chunlai Chen are the corresponding authors of this paper. Postdoctoral researcher Dr. Shouyue Zhang, PhD student Ao Sun, and PhD student Shuo Lin from the School of Life Sciences at Tsinghua University and PhD student Jingmei Qian from the Chinese Academy of Sciences are the co-first authors. In addition, the research was supported by Dr. Caixia Gao, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, and Dr. Mengqiu Dong, Beijing Institute of Life Sciences. The study received substantial support from the National Key Research and Development Program, the National Natural Science Foundation of China, the Ministry of Agriculture, the Scientific Exploration Award, and funding from Tsinghua University and Peking University.
Link to the paper: https://www.nature.com/articles/s41586-024-07486-x
Editor: Li Han

