Recently, Prof. Zhucheng Chen’s group at the School of Life Sciences of Tsinghua University made a breakthrough in the field of chromatin remodeling. This study reports the high-resolution cryo-electron microscope (cryoEM) structure of the human chromatin remodeling complex PBAF bound to a nucleosome in the active state, which provides mechanistic insights into PBAF assembly and regulation by various chromatin cues. Furthermore, this study provides the basis for the therapeutic implications of numerous BAF/PBAF-associated human diseases.
In eukaryotes, DNA wraps around histone octamer to form nucleosomes, the repeating unit of chromatin. The formation of chromatin is essential for a genome package. Yet the interaction between DNA and histones must be overcome in order to access the genetic materials for transcription, DNA replication, DNA damage repair and related factors. ATP-dependent chromatin remodeling complexes utilize the energy of ATP to slide, eject, and recompose nucleosomes, regulating chromatin accessibility and structure.
BAF (BRG1-associated factors) and PBAF (polybromo-associated BRG1-associated factors) chromatin remodeling complexes play a major role in the control of chromatin structure and gene expression, and are widely involved in the development and differentiation program in mammalian cells. In recent years, high-throughput DNA sequencing reveal that over 20% of human cancers are associated with mutations in the BAF and PBAF complex, making these complexes among the most frequently mutated chromatin factors in human cancers. Importantly, BAF and PBAF are considered potential drug targets for the treatment of these diseases. Although the BAF and PBAF complexes were identified as early as 1996, their assembly and the mechanisms of action remain unclear. Due to the advance in cryoEM technology, high-resolution structures of yeast SWI/SNF family complexes and human BAF complexes were reported in 2019 and 2020 respectively. However, the questions of PBAF complex assembly, the mechanism of nucleosomes recognition, and the functional difference between PBAF and BAF complexes remain to be investigated. In particular, the reported structure of the catalytic subunit (SMARCA4/BRG1) of the BAF complex was in the inactive state, and had a limited power to inform disease mechanisms.
Prof. Zhucheng Chen’s group has worked in the field of chromatin biology for a long time, and has published a series of important results on chromatin remodeling. In this PBAF project, the team reconstituted the PBAF complexes in vitro and used cryoEM to determine the high-resolution structure of PBAF bound to a nucleosome (3.4Å).
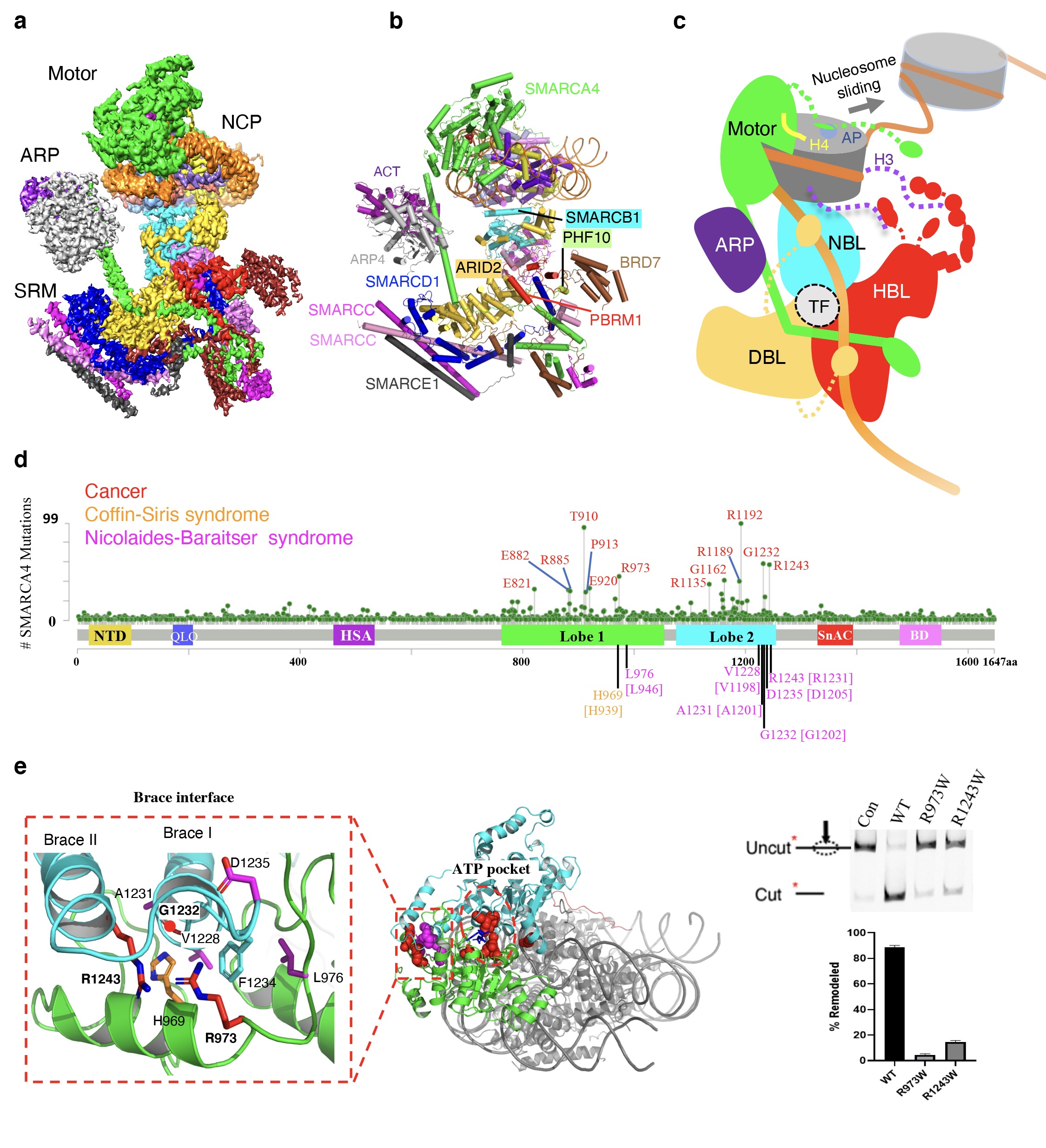
Structure of the human PBAF complex bound to a nucleosome and the implications of disease-associated mutations
(a) Composite cryoEM map of the PBAF-nucleosome complex. (b) Ribbon model of PBAF-nucleosome complex. The 12 subunits of PBAF complex are indicated. (c) Schematic model for nucleosome recognition and chromatin remodeling of PBAF complex. (d) Summary of 3703 missense mutations of SMARCA4 in the non-redundant cancer samples reported in COSMIC and cBioPortal (case numbers in parentheses) and in SMARCA2-associated Coffin-Siris syndrome and Nicolaides-Baraitser syndrome (the equivalent residues of SMARCA2 are indicated in the square brackets). Residue numbering is based on the canonical SMARCA4/2 isoforms. (e) Map of the cancer-associated hotspot mutations of SMARCA4 around ATP-binding pocket and Brace-helix interface. Local map shows the distribution of high-frequency mutations at the Brace-helix interface. The chromatin remodeling activities of wild type (WT) and two Brace-helix mutants are shown on the right. Compared with the WT complex, the R973W or R1243W mutant complexes displayed five-tenfold losses of the remodeling activity in vitro.
As shown by the structure, the 12 subunits of the PBAF complex are delineated into three functional modules: the catalytic motor module, the regulatory actin-related protein (ARP) module, and the substrate recruitment module (SRM) (Fig. a). The majority of the auxiliary subunits are assembled into the SRM module, which is responsible for the recognition of the nucleosome signals. Interestingly, the function-related subunits within the SRM cluster to form three lobe-like submodules, including the nucleosome-binding lobe (NBL), histone-tail binding lobe (HBL) and DNA-binding lobe (DBL) (Fig. b). In particular, three PBAF-specific subunits, PBRM1, PHF10 and BRD7, form the unique HBL, which contains six BDs and two BAH domains of PBRM1, two PHDs of PHF10, and one BD of BRD7. These domains gather at the HBL, and may collectively function as a super-H3-tail-binding unit for genome targeting of PBAF. The study also suggests possible mechanisms by which PBAF interacts with transcription factors (TFs) to regulate gene expression (Fig. c).
Notably, the motor subunit SMARCA4 is in the active state in the structure of PBAF. The active conformation of SMARCA4 clearly reveals that the disease-associated hotspot mutations are primarily located around the ATP-binding pocket and the Brace-helix interface. The Brace-helix interface was first discovered in the yeast chromatin remodeling protein Snf2 by Professor Chen's group in 2017. It forms across the two catalytic domains (Lobe1 and Lobe2) of the motor, and is crucial for coupling ATP hydrolysis to chromatin remodeling. In this work, the researchers found that multiple high-frequency mutations of SMARCA4 are located at the newly formed Brace-helix interface. For instance, Arg973, Gly1232 and Arg1243 are cancer-associated hotspot mutations, and they map to the Brace-helix interface (Fig. e). Furthermore, multiple missense mutations in the closely related paralog SMARCA2 found in patients with Nicolaides-Baraitser syndrome or Coffin-Siris syndrome also cluster at the Brace-helix interface (Fig. d). Biochemical experiments confirmed that mutations of these sites significantly reduced the chromatin remodeling activity in vitro, suggesting loss of function of the BAF/PBAF complex in patients. In addition, the high-quality EM density reveals the mechanism of nucleosome binding by three arginine anchors of the SnAc domain of SMARCA4. These arginine anchors are highly conserved in the Snf2-like enzymes, suggesting a common mechanism of nucleosome recognition.
In summary, this work not only elucidates the mechanisms of PBAF complex assembly and nucleosome recognition, but also provides insights into the regulation by various chromatin cues, and the impact of disease-related mutations. Furthermore, the study will pave the way for the development of drugs targeting the BAF/PBAF complex.
On April 27, the findings were published online in Nature, entitled "Structure of human PBAF chromatin remodeling complex bound to a nucleosome". Professor Zhucheng Chen from the School of Life Sciences of Tsinghua University is the corresponding author. Ph.D. student Junjie Yuan (School of Life Sciences /Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University) and Kangjing Chen (School of Life Sciences, Tsinghua University) are co-first authors of this work. Dr. Wenbo Zhang made important contributions to this research. This work was supported by the National Key Research and Development Program, the National Natural Science Foundation of China, the Advanced Innovation Center for Structural Biology, the Tsinghua-Peking Joint Center for Life Sciences and the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for the cryo-EM facility.
Original Article Link: https://www.nature.com/articles/s41586-022-04658-5
Editors: John Olbrich, Li Han

