Opioids such as morphine and fentanyl are potent analgesics widely used in clinical practice. They alleviate pain by activating the µ-opioid receptor (µOR) in the brain. With a history dating back over 5,000 years to ancient Sumerian civilization, opioids have long been valued for their pain-relieving properties. However, the long-term use of opioids can lead to dependence, increased tolerance, and severe side effects, including constipation and potentially fatal respiratory depression. Since 2000, opioid overdoses have resulted in approximately 400,000 deaths. In 2018 alone, the opioid crisis cost the U.S. around $700 billion, about 3.4% of its GDP. Understanding the activation mechanism of µOR and its interactions with different drugs is crucial for developing novel analgesics with fewer side effects.
On April 10th, a landmark paper titled "Ligand Efficacy Modulates Conformational Dynamics of the µ-Opioid Receptor" was published in the journal Nature. This research, led by Dr. Chunlai Chen from Tsinghua University, Dr. Brian Kobilka from Stanford University, and Dr. Wayne Hubbell from the University of California, combined Double Electron–Electron Resonance (DEER) and Single-Molecule Fluorescence Resonance Energy Transfer (smFRET) methods to explore the molecular basis of µOR activation and signal transduction.
The µ-opioid receptor (µOR) belongs to the G protein-coupled receptor (GPCR) family, a group of membrane proteins crucial for receiving external signals and initiating cellular responses. GPCRs play essential roles in various physiological processes, making them prime targets for drug development. GPCRs are known for their complex and dynamic nature, capable of adopting multiple conformations in response to ligands, which in turn activate downstream G proteins or β-arrestin pathways (Figure 1).
Traditional structural biology techniques like X-ray crystallography and cryo-electron microscopy (cryo-EM) typically capture only the most stable GPCR conformations. However, DEER can reveal different conformations and their populations with sub-angstrom resolution, while smFRET provides real-time conformational dynamics.
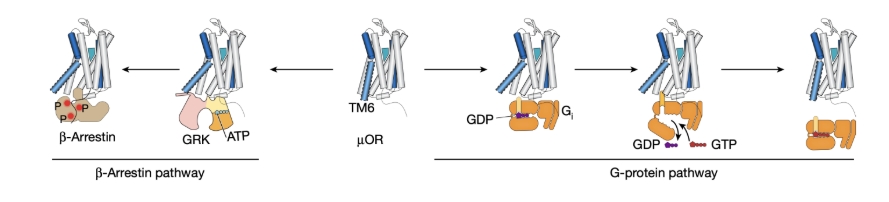
Figure 1 Two downstream signalling pathways of µOR.
Using DEER (Figure 2), the researchers identified four distinct µOR conformations: two inactive states (R1 and R2) and two active states (R3 and R4). Previous structural studies had only identified one inactive and one active conformation. High-efficacy ligands, such as lofentanil, significantly increased the proportion of µOR in active conformations, even in the absence of downstream G-protein binding. In contrast, low-efficacy ligands, like TRV130, had a lesser effect.
When G proteins were present, all ligands, regardless of efficacy, stabilized the R3 and R4 active conformations, with R3 being predominant. This suggests that ligand efficacy correlates with the ability to stabilize active conformations when alone. Moreover, β-arrestin predominantly bound to the R4 conformation, suggesting different binding conformations of µOR for G-protein and β-arrestin.
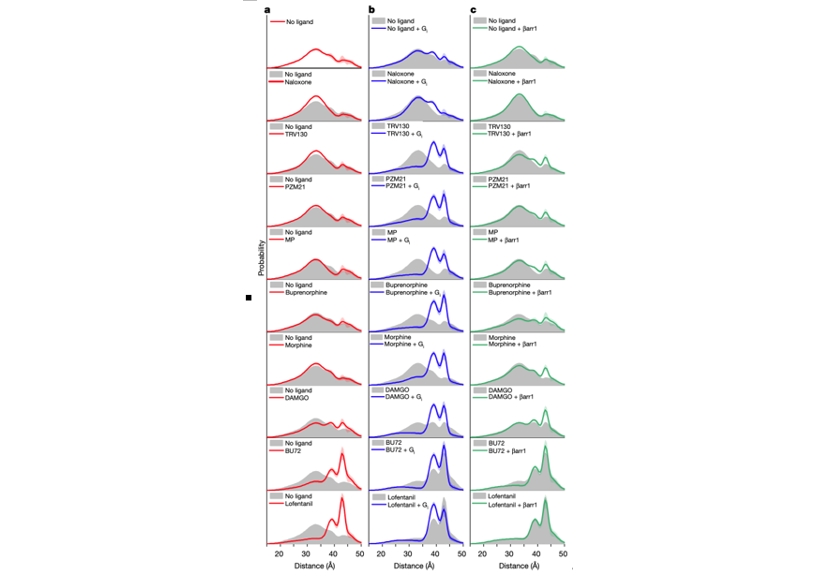 Figure2. Conformations of µOR revealed by DEER.
Figure2. Conformations of µOR revealed by DEER.
SmFRET experiments corroborated the DEER findings (Figure 3). µOR favored inactive conformations with low-efficacy ligands and active conformations with high-efficacy ligands. Labeling the intracellular loop 2 (ICL2) instead of TM4, smFRET detected two high-FRET states in the presence of antagonists or low-efficacy ligands, indicating conformational changes related to ligand efficacy. Additionally, smFRET observed µOR conformational transitions between active and inactive states in the presence of G protein and varying GDP concentrations (Figure 4), revealing two G protein-bound states: GDP-bound (EFRET = 0.6) and GDP-free (EFRET = 0.5). High-efficacy ligands accelerated G protein binding and GDP release, enhancing GTP binding and downstream signaling.
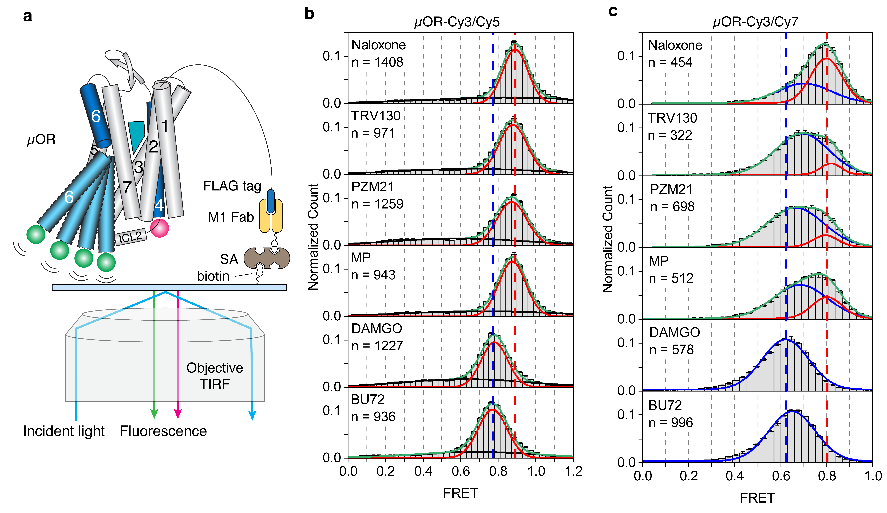
Figure3. SmFRET distributions of µOR–Cy3/Cy5 (b) and µOR–Cy3/Cy7 (c) in the presence of different ligands.
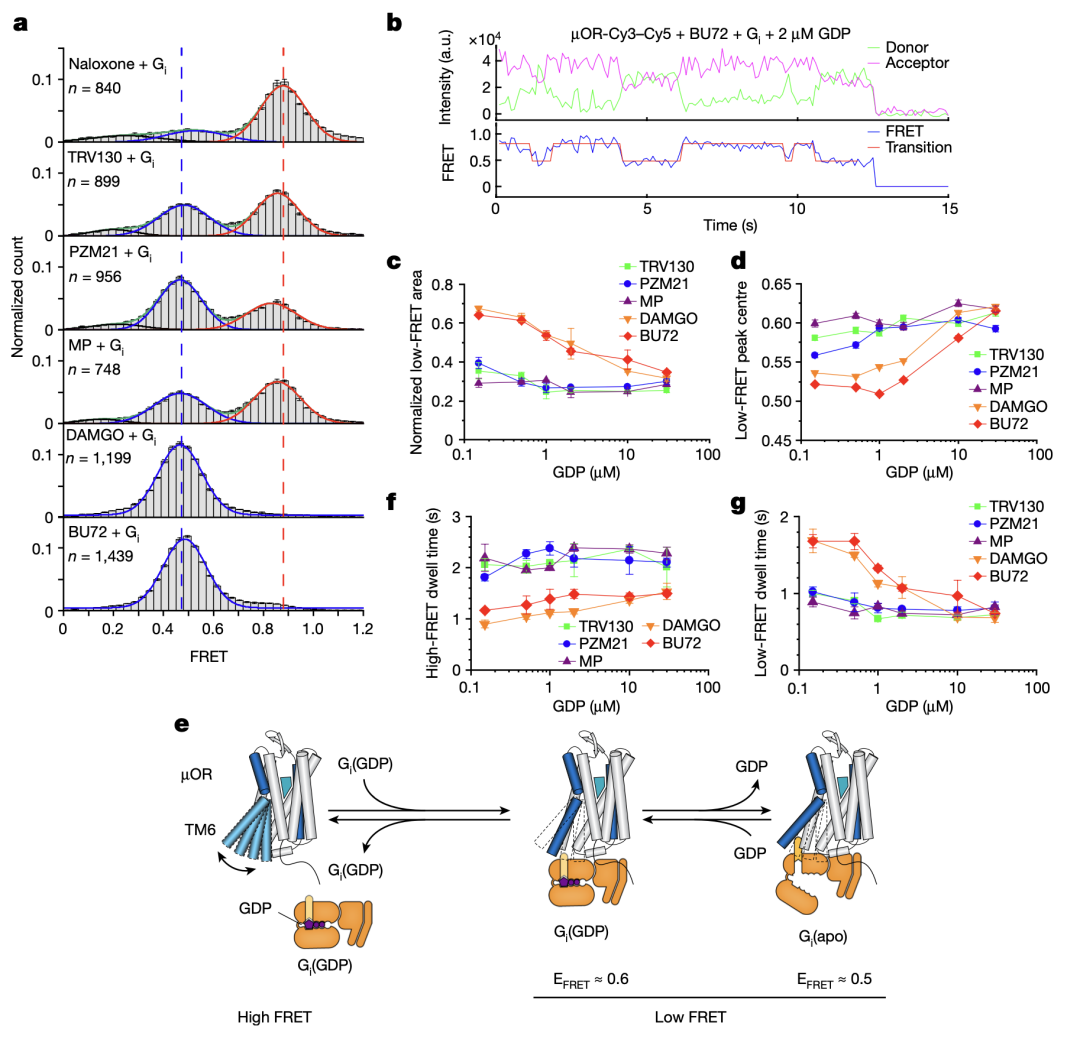
Figure 4. Structural dynamics of the µOR revealed by smFRET.
This study not only advances our understanding of µOR activation mechanisms but also provides a molecular foundation for developing novel analgesic drugs with reduced side effects. Further insights into GPCR dynamics and activation will guide the creation of safer and more effective pain management therapies.
Drs. Chunlai Chen, Brian Kobilka, and Matthias Elgeti are co-corresponding authors, and Drs. Jiawei Zhao and Matthias Elgeti are co-first authors. The study was supported by several funding sources, including the National Natural Science Foundation of China, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, State Key Laboratory of Membrane Biology, and Tsinghua–Peking Joint Center for Life Sciences.
Link to the paper:
https://www.nature.com/articles/s41586-024-07295-2
Editor:Li Han

